Abstract
Background: Intestinal microbiome constitutes a symbiotic ecosystem that is essential for health, and changes in its composition/function cause various illnesses. Biochemical milieu shapes the structure and function of the microbiome. Recently, we found marked differences in the abundance of numerous bacterial taxa between ESRD and healthy individuals. Influx of urea and uric acid and dietary restriction of fruits and vegetables to prevent hyperkalemia alter ESRD patients' intestinal milieu. We hypothesized that relative abundances of bacteria possessing urease, uricase, and p-cresol- and indole-producing enzymes is increased, while abundance of bacteria containing enzymes converting dietary fiber to short-chain fatty acids (SCFA) is reduced in ESRD. Methods: Reference sets of bacteria containing genes of interest were compiled to family, and sets of intestinal bacterial families showing differential abundances between 12 healthy and 24 ESRD individuals enrolled in our original study were compiled. Overlap between sets was assessed using hypergeometric distribution tests. Results: Among 19 microbial families that were dominant in ESRD patients, 12 possessed urease, 5 possessed uricase, and 4 possessed indole and p-cresol-forming enzymes. Among 4 microbial families that were diminished in ESRD patients, 2 possessed butyrate-forming enzymes. Probabilities of these overlapping distributions were <0.05. Conclusions: ESRD patients exhibited significant expansion of bacterial families possessing urease, uricase, and indole and p-cresol forming enzymes, and contraction of families possessing butyrate-forming enzymes. Given the deleterious effects of indoxyl sulfate, p-cresol sulfate, and urea-derived ammonia, and beneficial actions of SCFA, these changes in intestinal microbial metabolism contribute to uremic toxicity and inflammation.
Introduction
The enormous community of microbes that resides in the intestinal tract (gut microbiome) constitutes a symbiotic ecosystem that provides protection against invasion by pathogens and contributes to nutritional balance by facilitating absorption of complex carbohydrates and production of amino acids and various micronutrients. The integrity and proper function of the gut microbiome is essential for health, and alteration in the composition or function of the microbiome is associated with various illnesses, such as inflammatory bowel disease, diabetes, cardiovascular diseases, cancer, obesity, chronic inflammation, dyslipidemia, and allergic disorders. In a recent study [1], we found marked differences in the abundance of numerous bacterial taxa recovered in the feces between end-stage renal disease patients and healthy control individuals. By disturbing the normal symbiotic relationship with the host, the observed changes in the composition of the intestinal microbiome may have adverse consequences. For instance, changes in the composition of the microbiome may contribute to uremic toxicity, inflammation, and various other complications of chronic kidney disease (CKD) by promoting generation of toxic products and lowering the protective functions and beneficial by-products of the normal flora.
The biochemical milieu plays a decisive role in shaping the composition and function of the gut microbiome. Renal failure markedly alters the biochemical environment of the gastrointestinal tract. For instance, accumulation of urea in the body fluids of renal failure patients leads to its heavy influx into the gastrointestinal tract where it is hydrolyzed by microbial urease to produce large quantities of ammonia [CO(NH2)2 + H2O→>CO2 + 2NH3] [2,3]. Ammonia is then converted to ammonium hydroxide [NH3 + H2O→NH4OH], which elevates the gut's luminal pH and causes mucosal damage and irritation [4,5]. In addition, significant amounts of uric acid and oxalate are secreted in the colon in an adaptive response to the loss of their renal excretion [6,7,8,9]. Because of the influx of massive quantities of urea and uric acid in the gut of ESRD patients, the altered milieu may favor microbes capable of utilizing these substrates.
Another important factor affecting the intestinal milieu of ESRD patients is dietary restriction of fruits and vegetables, commonly imposed to limit potassium intake and prevent hyperkalemia. The inevitable reduction of dietary fiber content can potentially lead to reduced populations of the bacterial families capable of utilizing fiber, which is the primary substrate for normal intestinal microbiota. Additionally, several studies have documented increased blood concentrations of indoxyl sulfate and p-cresol sulfate [10] in ESRD patients, and sulfur-containing fecal volatile compounds such as dimethyldisulfide, dimethyltrisulfide, ethanethioic acid S methyl ester, and butanethioic acid S-methyl ester in CKD animals [10,11,12]. Tryptophanase and hydroxyphenylacetate decarboxylase mediate the initial steps in production of indoxyl sulfate and p-cresol sulfate (from tryptophan and tyrosine) [13,14,15,16], whose serum levels markedly increase due to reduced clearance in advanced CKD. Finally, use of iron-containing or resin-based phosphate binders can potentially impact the gut microbiome since the former increases the availability of iron which is essential for microbial growth, while by binding the bile acids the latter products reduce the availability of bile acids which are major substrates for certain microbial species. Based on the above observations, we predicted that the heavy influx of urea and uric acid into the intestinal tract in patients with chronic renal failure may accommodate expansion of microbial families possessing urease and uricase, enabling them to utilize these compounds as substrates. We further predicted that increased serum levels of indoxyl sulfate and p-cresol sulfate may signify the relative prominence of microbes possessing enzymes capable of generating indoxyl and p-cresol. In contrast, we considered that dietary fiber restriction may cause contraction of normally prominent bacterial families possessing fermenting enzymes like butyrate kinase. The present in silico study was undertaken to test these hypotheses.
Methods
Patients and Controls
As reported in greater detail elsewhere [1], 24 stable ESRD patients (6 men and 18 women, aged 57 ± 14 years) maintained on hemodialysis and 12 normal subjects (4 men and 8 women, aged 51 ± 12 years) were recruited for this study. The underlying causes of ESRD in the study population included diabetic nephropathy, hypertensive nephrosclerosis, polycystic kidney disease, lithium nephropathy, and glomerulonephritis. Hemodialysis blood access consisted of arteriovenous fistulas, polytetrafluoroethylene grafts, or tunneled dual lumen central catheters. All but 4 patients were taking darbopoetin, and all but 2 were taking a phosphate binder. All patients followed a potassium-restricted diet. Individuals with a history of acute illness or antibiotic therapy during the prior 3 months were excluded. The study protocol was approved by Human Subjects Institutional Review Board of the University of California Irvine (HS# 2007-5572) and completed with the assistance of the University of California Irvine Institute of Clinical Translational Science.
Prominent Intestinal Bacterial Families in ESRD and Control Groups
Bacteria differing in relative abundances between ESRD patients' and control participants' fecal samples [1] were grouped by family for analysis. Note: Several operational taxonomic units (OTUs) were reannotated to reflect the latest Greengenes taxonomy [17] and to classify the previously unclassified OTUs.
Bacterial Families Containing Enzymes of Interest
Records of all reported bacteria that possess the genes encoding urease, uricase, tryptophanase, phosphotransbutyrylase, or butyrate kinase were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG). KEGG is an online repository containing gene catalogs from sequenced genomes as well as information related to higher-order cellular and enzymatic pathways. The KEGG database was searched for the corresponding KEGG Orthology (KO) identifier(s) for each enzyme of interest. The KO ids were then used to obtain lists of gene orthologs pertaining to specific genomes, which later were filtered to contain only unique lineages of bacteria that possess the enzyme of interest. The resulting collection of unique bacterial families from each query was considered to be the primary reference set. From this reference set, a subset of bacteria possessing a particular enzyme and having higher relative abundances in either ESRD or control samples was identified.
For p-cresol, the reference set of bacterial families used for this study was compiled based on the bacterial species reported in the literature to have the ability to produce p-cresol from tyrosine (Clostridium difficile [16,18,19,20]) or contribute to p-cresol formation either by converting tyrosine to p-hydroxyphenylacetate (pHPA; e.g. various clostridia or Proteus vulgaris [15]) or converting pHPA to p-cresol (e.g. Lactobacillaceae [14]). This literature review for bacteria of interest was used instead of a KEGG search because a key enzyme responsible for the transamination of L-tyrosine to p-hydroxyphenylacetic acid (pHPAA) has not been named officially in the literature, and gene orthologs of organisms capable of performing decarboxylation of pHPAA to p-cresol (i.e. having para-hydroxyphenylacetate decarboxylase; online suppl. table S5; for all online suppl. material, see www.karger.com/doi/10.1159/000360010) were not all available in KEGG.
Statistical Analyses
The lists of families differing in relative abundance between ESRD patients and control samples were used in a test along with the lists of bacterial families identified to have the enzymes of interest. The likelihood of bacterial families being in both lists by chance was assessed using hypergeometric distributions and the associated probability function. Overrepresentation occurs when there is a larger than expected subset of bacterial families possessing a particular enzymatic function out of the total set of highly abundant bacterial families. The distribution and the probability function allowed us to analyze the representation of a specific bacterial family in our study and determine whether or not this representation occurred due to chance. Additional details are provided in the online supplementary material.
Results
Bacterial families containing taxa differing between ESRD patients' and control participants' samples are presented in table 1 (with updated taxonomies presented in online suppl. table S1). Detailed information is included in the online supplementary material.
Urease and Uricase
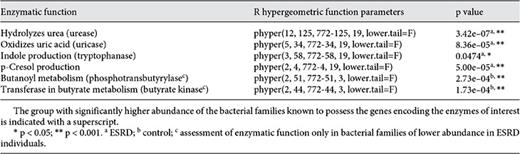
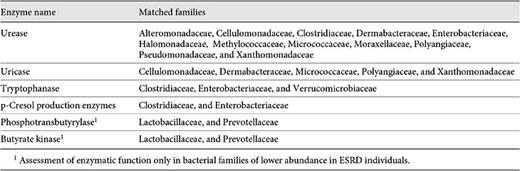
One hundred and twenty-five bacterial families contained the functional gene urease according to the KEGG database (online suppl. table S2; see table 2 for KO identifiers). Twelve of the 19 microbial families (63%) with greater abundances in ESRD patients were among the urease-possessing families (table 3). These 12 families were: Alteromonadaceae, Cellulomonadaceae, Clostridiaceae, Dermabacteraceae, Enterobacteriaceae, Halomonadaceae, Methylococcaceae, Micrococcaceae, Moraxellaceae, Polyangiaceae, Pseudomonadaceae, and Xanthomonadaceae. Similarly, 34 distinct bacterial families considered to have the uricase gene were identified from KEGG (online suppl. table S3). Among the 19 bacterial families that were most abundant in the ESRD patients, 5 families (26%) had members that possessed the uricase gene. These included Cellulomonadaceae, Dermabacteraceae, Micrococcaceae, Polyangiaceae, and Xanthomonadaceae families (table 3).
List of KEGG orthology identifiers that were used in querying the database for records of bacteria with specific genes of interest (does not include enzymes for paracresol production)
Bacterial families with members containing urease, uricase, tryptophanase, and p-cresol-producing enzymes which were more abundant in the ESRD group, and the bacterial families with members possessing phosphotransbutyrylase and butyrate kinase which were less abundant in ESRD patients compared to healthy control group
Tryptophanase and p-Cresol Formation
There were 58 families found in KEGG containing the tryptophanase gene (online suppl. table S4). Among the 19 bacterial families that were more abundant in the ESRD patients, 3 families had members that possessed the tryptophanase gene. These included Clostridiaceae, Enterobacteriaceae, and Verrucomicrobiaceae. Additionally, 4 families of bacteria capable of deaminating tyrosine into p-cresol were found from the literature review (online suppl. table S5). Clostridiaceae and Enterobacteriaceae were the two families of that reference set that also were part of the 19 microbial families with increased relative abundance in ESRD patients.
Butyrate-Producing Enzymes
Fifty-one bacterial families in KEGG contained phosphotransbutyrylase (online suppl. table S6). Two families, Lactobacillaceae and Prevotellaceae, of the 3 that had lower abundance in ESRD samples were among those 51 families. Similarly, the KEGG repository contained 44 bacterial families with the butyrate kinase gene (online suppl. table S7). Lactobacillaceae and Prevotellaceae, which were among the families with reduced abundance in the ESRD samples, include members that possess the butyrate kinase gene.
Statistical Tests
Hypergeometric distribution tests provided probabilities of bacterial families meeting two criteria by chance alone: (1) having elevated (or reduced) relative abundances in ESRD patients, and (2) having the metabolic genes of interest. The probability of having 63% urease-possessing bacterial families in ESRD patients versus control subjects due to random chance was evaluated, and a p value <0.001 was obtained (table 4). Likewise, the probability tests for uricase (26%), p-cresol production (10%), phosphotransbutyrylase (67%), and butyrate kinase (67%) all yielded p values <0.001. The p value was <0.05 when the probability of having 16% (3 out of 19) bacterial families containing tryptophanase-encoding genes was examined.
Discussion
The in silico tests reported here were performed to determine if the phylogenetic data could support the hypotheses of increased or decreased relative abundance of bacterial families containing genes encoding enzymes predicted to be important in ESRD patients compared to the healthy controls. Selection pressures on the part of hosts and microbes shape the structure, composition, and function of the gut microbiota. As noted earlier, advanced renal failure results in profound alteration of the biochemical milieu of the gastrointestinal tract by several mechanisms, including massive influx of urea and secretion of uric acid into the intestinal tract. Hydrolysis of urea by microbial urease [3] in the gut forms a large quantity of ammonia, which is rapidly converted to ammonium hydroxide. Generation of these products plays a key role in the development of uremic enterocolitis [5,6]. In addition, recent studies have demonstrated the central role of urea-derived NH3 and ammonium hydroxide in the breakdown of the gut's epithelial barrier structure and function and its contribution to systemic inflammation [21,22]. There is mounting evidence supporting the presence of the gastrointestinal barrier dysfunction and its role in the pathogenesis of systemic inflammation in uremic humans and animals [23]. ESRD patients commonly exhibit histological evidence of chronic inflammation throughout the gastrointestinal tract [6]. Moreover endotoxemia is invariably present in ESRD patients in the absence of clinical infection [24,25,26]. Integrity of the tight junction, which seals the gap between the epithelial cells of the gastrointestinal tract, is critical in preventing the entry of the microbial toxins, antigens, and other harmful products in the subepithelial tissues and the internal milieu. By enabling the absorption of these products, the intestinal epithelial barrier impairment leads to local and systemic inflammation, which are invariably present in ESRD patients and play a major role in the pathogenesis of cardiovascular disease, anemia, protein energy wasting and various other complications [27,28,29]. These observations point to increased intestinal permeability and barrier dysfunction in this population. However, until recently the mechanism by which uremia increases intestinal epithelial permeability was not known. In a recent study, we found heavy losses of the key protein constituents of colonic epithelial tight junction in rats with CKD [30,31], which unraveled the source of uremia-associated endotoxemia. In a series of in vitro studies, we identified urea and the by-products of its hydrolysis by microbial urease (i.e. ammonia and ammonium hydroxide) as the major mediator of uremia-induced disruption of the intestinal barrier function and structure [21]. Together, these observations illustrate the critical role of urea and urease-containing microbial families in the pathogenesis of inflammation and the associated complications in patients with CKD. Moreover, the influx of urea and its conversion to ammonia alter the intestinal pH, which is known to play a major role in shaping other microbial communities [32,33,34] and to affect production of metabolites by the gut bacteria [13]. The modification of luminal pH also may contribute to the expansion of bacteria that thrive at higher pH.
The results of the present study support the hypothesis that chronic renal failure results in expansion of bacterial families possessing urease. This observation is supported by earlier studies by Brown et al. [35] who found an increase in both fecal urease activity and culturable urease positive bacteria in uremic patients. In fact, relative increases in members of the Enterobacteriaceae, Pseudomonadaceae, and Clostridiaceae were found in both studies.
Unlike urea and creatinine whose serum concentrations rise dramatically, serum uric acid concentration increases only minimally despite loss of its renal excretion in patients with advance renal failure. To explore the reason for this disparity, in a series of earlier studies we found active secretion of uric acid by colonic epithelium in the uremic animals [7,8] which accounts for the lack of significant hyperuricemia in chronic renal failure. The finding of relative expansion of uricase-possessing bacterial families in our ESRD patients represents an adaptive response on the part of the gut microbiome to the availability of this substrate.
The effect of uremia is compounded by strict dietary restrictions imposed to prevent hyperkalemia by limiting consumption of potassium-rich fruits, vegetables, and high-fiber products. These foods contain most of the normal dietary indigestible complex carbohydrates that serve as the primary source of nutrients for the gut microbiota. Fermentation of these complex carbohydrates by bacteria in the intestine leads to formation of short-chain fatty acids (SCFA), such as butyrate, as well as volatile compounds, such as methane and methanol. In fact, in a recent study of the ESRD patients, Lee et al. [36] found a significant reduction in the production of methanol, which is a by-product of the fermentation of un-absorbable complex carbohydrates. The SCFA, particularly butyrate, produced by the bacteria are the major source of energy for colonocytes. The dependence of colonocytes on butyrate produced by bacteria was demonstrated in an elegant study by Donohoe et al. [37] who showed ATP depletion and autophagy in colonocytes of the germ-free mice and their reversal by addition of butyrate. We hypothesized that limitation of fiber in ESRD patients' diet would lead to a contraction of the SCFA-producing families that ferment complex carbohydrates. Prevotellaceae have been associated with fiber-rich diets, especially the plant-derived polysaccharides [38]. The updated taxonomy resulted in two bacterial taxa being classified as Prevotellaceae. These bacteria were less abundant in the ESRD patients than in controls, which is consistent with our predicted reduction of butyrate-producing and associated bacteria in ESRD patients occasioned dietary restrictions.
Reduced dietary fiber intake in ESRD patients coupled with the observed reduction in the population of butyrate-forming bacteria may contribute to the pathogenesis of inflammation in this population. Systemic inflammation in ESRD patients is mediated by activation of innate immune system, orchestrated by monocytes, macrophages, granulocytes and cellular constituents of other organs/tissues [39]. This is compounded by the depletion and dysfunction of regulatory T lymphocytes, which play a central role in maintaining immunological self-tolerance, limiting the inflammatory response to foreign antigens, and cessation of the T cell-mediated immunity upon completion of the immune reaction [39,40]. T regulatory cell depletion and dysfunction plays a major role in the pathogenesis of systemic inflammation and its adverse consequences in patients with CKD. In a recent study, Smith et al. [41] showed that the SCFA (propionate, acetate, and butyrate) selectively expand the pool of the regulatory T cells in the large intestine. Expansion of the regulatory T cells by these microbial products helps to attenuate inflammation by suppressing the activity of inflammatory cells. The role of dietary fiber is further supported by the work of Krishnamurthy et al. [42], who found a link between low-fiber diets and both increased inflammation and mortality in a CKD population. Together, these findings have unraveled a new link between bacterial products and a major anti-inflammatory pathway in the gut.
Serum concentrations of the uremic toxins indoxyl sulfate and p-cresol sulfate are markedly elevated in patients with ESRD [12,43]. Using control and germ-free mice, Wikoff et al. [44] demonstrated the role of bacteria as the initiators of p-cresol sulfate, indoxyl sulfate, and several other compounds that accumulate in uremia. Tryptophanase and hydroxyphenylacetate decarboxylase catalyze the formation of indole from tryptophan and p-cresol from tyrosine, respectively, which are absorbed and sulfated by the liver [12,13]. The present study revealed significant expansion of two tryptophanase-containing bacterial families (Clostridiaceae and Enterobacteriaceae) in the ESRD patients. This could lead to increased indoxyl sulfate production, which together with the lack of renal excretion may account for its elevated plasma concentration in this population. p-Cresol formation (e.g. from tyrosine by C. difficile [18,20] and from pHPAA by some lactobacilli [14,15]) with reduced clearance of the sulfated molecule also contributes to uremic toxicity. The results of the present study are consistent with the hypothesis that the relative abundance of p-cresol-producing bacteria is significantly greater in ESRD patients than in healthy subjects. The results of earlier studies by Smith and Macfarlane [13,45] suggest that the reduction of fermentable complex carbohydrates and elevated colonic pH may contribute to the increased production of phenolic and indolic compounds by colonic bacteria. Using batch fecal culture fermentation studies, they showed the net production of phenolic compounds by mixed populations of intestinal bacteria was reduced by approximately 33% at pH 5.5 compared to pH 6.8, and by 60% in the presence of a fermentable carbohydrate. These findings highlight the possible contribution of reduced dietary fiber and elevation of colonic pH by urea-derived ammonia to increased levels of indoxyl sulfate and p-cresol sulfate in ESRD patients.
In conclusion, patients with ESRD exhibited significant expansion of bacterial families possessing urease, uricase, and indole- and p-cresol-forming enzymes and reduction of bacterial families possessing butyrate-forming enzymes. In view of the damaging effects of the urease-derived ammonia on the intestinal epithelial barrier, the toxic effects of indoxyl sulfate and p-cresol sulfate, and the protective effect of the microbially derived SCFA, the observed changes in the intestinal microbiota likely contribute to the uremic toxicity and local and systemic inflammation in ESRD population. Future studies are planned to investigate the gut microbial metagenome, metatranscriptome, metabolome, and host transcriptome in ESRD populations and explore the impact of dietary interventions aimed at mitigating the effect of uremia on intestinal microbiota.
Acknowledgements
The authors thank Jun Yuan and Zhenmin Ni for their contributions to the work underlying this article's PhyloChip data as presented in Vaziri et al. [1].
Disclosure Statement
The authors have no conflict of interest to declare. This project was, in part, supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR000153.
References
J. Wong and Y.M. Piceno contributed equally to this work.